Stochastic Calculus#
Deterministic dynamical systems#
Numerical solution of dynamical systems: inflation targetting#
import numpy as np
import matplotlib.pyplot as plt
def simulate_inflation(theta, pi_hat, pi_t0, Delta, N):
# Time grid
time = np.linspace(0, N * Delta, N + 1)
# Arrays to store the results
pi_forward = np.zeros(N + 1)
pi_backward = np.zeros(N + 1)
pi_analytical = np.zeros(N + 1)
# Initial condition
pi_forward[0] = pi_t0
pi_backward[0] = pi_t0
# Analytical Solution
pi_analytical = pi_hat + (pi_t0 - pi_hat) * np.exp(-theta * time)
# Forward Euler Scheme
for i in range(N):
pi_forward[i + 1] = pi_forward[i] + Delta * theta * (pi_hat - pi_forward[i])
# Backward Euler Scheme
for i in range(N):
pi_backward[i + 1] = (pi_backward[i] + Delta * theta * pi_hat) / (1 + Delta * theta)
return time, pi_forward, pi_backward, pi_analytical
# Parameters for normal regime
theta_normal = 0.1
Delta_normal = 0.1
pi_hat = 2.0
pi_t0 = 5.0
N = 100
# Parameters for challenging regime
theta_challenging = 10.0
Delta_challenging = 0.1
# Simulate for normal regime
time_normal, pi_forward_normal, pi_backward_normal, pi_analytical_normal = simulate_inflation(
theta_normal, pi_hat, pi_t0, Delta_normal, N)
# Simulate for challenging regime
time_challenging, pi_forward_challenging, pi_backward_challenging, pi_analytical_challenging = simulate_inflation(
theta_challenging, pi_hat, pi_t0, Delta_challenging, N)
# Plot the results for normal regime
plt.figure(figsize=(12, 8))
plt.plot(time_normal, pi_forward_normal, label='Forward Euler (Normal)', marker='o', linestyle='--')
plt.plot(time_normal, pi_backward_normal, label='Backward Euler (Normal)', marker='x', linestyle='--')
plt.plot(time_normal, pi_analytical_normal, label='Analytical Solution (Normal)', linestyle='-', linewidth=2)
plt.xlabel('Time')
plt.ylabel('Inflation Rate')
plt.title('Inflation Targeting Model Simulation (Normal Regime)')
plt.legend()
plt.grid(True)
plt.show()
# Plot the results for challenging regime
plt.figure(figsize=(12, 8))
plt.plot(time_challenging, pi_forward_challenging, label='Forward Euler (Challenging)', marker='o', linestyle='--')
plt.plot(time_challenging, pi_backward_challenging, label='Backward Euler (Challenging)', marker='x', linestyle='--')
plt.plot(time_challenging, pi_analytical_challenging, label='Analytical Solution (Challenging)', linestyle='-', linewidth=2)
plt.xlabel('Time')
plt.ylabel('Inflation Rate')
plt.title('Inflation Targeting Model Simulation (Challenging Regime)')
plt.legend()
plt.grid(True)
plt.show()
The Wiener Process#
Simulation of the Wiener process (univariate)#
import matplotlib.pyplot as plt
import numpy as np
# Set the parameters for the Wiener process
T = 1.0 # total time
n = 1000 # number of steps
dt = T / n # time increment
t = np.linspace(0, T, n+1) # time points
# Number of paths to simulate
num_paths = 5
# Set up the plot
plt.figure(figsize=(10, 6))
# Simulate multiple paths
for _ in range(num_paths):
W = np.zeros(n+1) # Initialize the Wiener process for each path
for i in range(n):
W[i+1] = W[i] + np.sqrt(dt) * np.random.randn()
plt.plot(t, W, label=f'Path {_+1}')
# Customize the plot
plt.title('Sample Paths of the Wiener Process')
plt.xlabel('Time t')
plt.ylabel('Wiener Process W(t)')
plt.grid(True)
# Show the plot
plt.show()
Simulation using Gaussian processes#
import GPy
import numpy as np
import matplotlib.pyplot as plt
# Time points at which to sample the Wiener process
t = np.linspace(0, 1, 1000).reshape(-1, 1)
# Define the Brownian motion kernel
brownian_kernel = GPy.kern.Brownian(input_dim=1, variance=1.0)
# Create a GP model with zero mean function
mean_function = GPy.mappings.Constant(input_dim=1, output_dim=1, value=0)
model = GPy.models.GPRegression(t, np.zeros_like(t), kernel=brownian_kernel, mean_function=mean_function)
# Ensure correct model optimization
model.optimize()
# Sample paths from the GP model
num_samples = 5
samples = model.posterior_samples_f(t, size=num_samples)
# Plot the sampled paths
plt.figure(figsize=(10, 6))
for i in range(num_samples):
plt.plot(t, samples[:, :, i], label=f'Sample {i+1}')
plt.title('Sampled Paths from a Wiener Process using Gaussian Processes')
plt.xlabel('Time')
plt.ylabel('W(t)')
plt.show()
Brownian motion with drift#
Simulation using discrete paths#
import matplotlib.pyplot as plt
import numpy as np
# Set the parameters for Brownian motion with drift and volatility
T = 1.0 # total time
n = 1000 # number of steps
dt = T / n # time increment
t = np.linspace(0, T, n+1) # time points
# Brownian motion parameters
mu = 5 # drift
sigma = 2.0 # volatility
# Number of paths to simulate
num_paths = 5
# Set up the plot
plt.figure(figsize=(10, 6))
# Simulate multiple paths
for _ in range(num_paths):
X = np.zeros(n+1) # Initialize the process for each path
for i in range(n):
X[i+1] = X[i] + mu * dt + sigma * np.sqrt(dt) * np.random.randn()
plt.plot(t, X, label=f'Path {_+1}')
# Customize the plot
plt.title('Sample Paths of Brownian Motion with drift')
plt.xlabel('Time t')
plt.ylabel('X(t)')
plt.grid(True)
# Show the plot
plt.show()
Simulation using Gaussian Processes#
import GPy
import numpy as np
import matplotlib.pyplot as plt
# Parameters for Brownian motion with drift
S0 = 0.0 # Initial value
mu = 0.5 # Drift coefficient
sigma = 0.3 # Volatility
T = 1.0 # Total time
n = 1000 # Number of steps
t = np.linspace(0, T, n+1).reshape(-1, 1) # Time points
# Define the custom kernel for Brownian motion
class BrownianMotionKernel(GPy.kern.Kern):
def __init__(self, input_dim, sigma=1.0, active_dims=None):
super(BrownianMotionKernel, self).__init__(input_dim, active_dims, 'Brownian')
self.sigma = GPy.core.Param('sigma', sigma)
self.link_parameters(self.sigma)
def K(self, X, X2=None):
if X2 is None:
X2 = X
t1 = X
t2 = X2.T
# Covariance function of Brownian motion: sigma^2 * min(s, t)
cov_matrix = self.sigma**2 * np.minimum(t1, t2)
return cov_matrix
def Kdiag(self, X):
return self.sigma**2 * X[:, 0]
# No-op for gradient update since we are not optimizing the kernel
def update_gradients_full(self, dL_dK, X, X2=None):
pass
# No-op for kernel gradient since we are not optimizing the kernel
def gradients_X(self, dL_dK, X, X2=None):
return np.zeros(X.shape)
# Custom mean function to include drift
class BrownianMotionMean(GPy.core.Mapping):
def __init__(self, input_dim, output_dim, S0=0.0, mu=0.0):
super(BrownianMotionMean, self).__init__(input_dim, output_dim)
self.S0 = S0 # Initial value
self.mu = mu # Drift coefficient
def f(self, X):
# Mean function: S0 + mu * t
return self.S0 + self.mu * X
def update_gradients(self, dL_df, X):
pass
def gradients_X(self, dL_df, X):
return np.zeros(X.shape)
# Instantiate the Brownian motion kernel with parameters
bm_kernel = BrownianMotionKernel(input_dim=1, sigma=sigma)
# Instantiate the mean function with S0 and mu
mean_function = BrownianMotionMean(input_dim=1, output_dim=1, S0=S0, mu=mu)
# Provide the initial observation at t=0
t_data = np.array([[0.0]])
Y_data = np.array([[S0]])
# Create a GP model with the custom mean function and Brownian motion kernel
model = GPy.models.GPRegression(t_data, Y_data, kernel=bm_kernel, mean_function=mean_function)
# Set noise variance to a negligible value since we're simulating without observation noise
model.likelihood.variance.fix(1e-10)
# Sample paths from the GP model
num_samples = 5
samples = model.posterior_samples_f(t, size=num_samples)
# Plot the sampled paths
plt.figure(figsize=(10, 6))
for i in range(num_samples):
plt.plot(t, samples[:, :, i], label=f'Path {i+1}')
plt.title(f'Sampled Paths from Brownian Motion with Drift')
plt.xlabel('Time t')
plt.ylabel('S(t)')
plt.grid(True)
plt.legend()
plt.show()
Arithmetic Average of the Brownian Motion#
Simulation using discrete paths#
import matplotlib.pyplot as plt
import numpy as np
# Set the parameters for Brownian motion with drift and volatility
T = 1.0 # total time
n = 1000 # number of steps
dt = T / n # time increment
t = np.linspace(dt, T, n) # time points starting from dt to avoid division by zero
# Brownian motion parameters
S0 = 0.0 # initial value of S_u
mu = 0.5 # drift coefficient
sigma = 0.3 # volatility
# Number of paths to simulate
num_paths = 5
# Initialize array to store M_t for each path
M_t_paths = np.zeros((n, num_paths))
# Set up the plot
plt.figure(figsize=(12, 6))
# Simulate multiple paths
for path in range(num_paths):
# Initialize S_u for each path
S = np.zeros(n + 1) # n+1 because we include S0
S[0] = S0
# Simulate S_u using Euler-Maruyama method
for i in range(n):
dW = np.sqrt(dt) * np.random.randn() # Brownian increment
S[i + 1] = S[i] + mu * dt + sigma * dW
# Compute cumulative sum to approximate the integral
cumulative_S = np.cumsum(S[:-1]) * dt # Exclude the last S[n+1]
# Compute M_t at each time point
M_t = cumulative_S / t
M_t_paths[:, path] = M_t
# Plot the path
plt.plot(t, M_t, label=f'Path {path + 1}')
# Customize the plot
plt.title('Sample Paths of the arithmetic mean of a Brownian motion')
plt.xlabel('Time t')
plt.ylabel('M(t)')
plt.grid(True)
plt.legend()
# Show the plot
plt.show()
Simulation using Gaussian Processes#
import GPy
import numpy as np
import matplotlib.pyplot as plt
# Parameters for Brownian motion with drift
S0 = 0.0 # Initial value
mu = 0.5 # Drift coefficient
sigma = 0.3 # Volatility
T = 1.0 # Total time
n = 1000 # Number of steps
t = np.linspace(1e-6, T, n+1).reshape(-1, 1) # Time points, avoid t=0 to prevent division by zero
# Define the custom kernel based on the provided covariance function
class TimeAverageKernel(GPy.kern.Kern):
def __init__(self, input_dim, sigma=1.0, active_dims=None):
super(TimeAverageKernel, self).__init__(input_dim, active_dims, 'TimeAverage')
self.sigma = GPy.core.Param('sigma', sigma)
self.link_parameters(self.sigma)
def K(self, X, X2=None):
if X2 is None:
X2 = X
t1 = X[:, 0].reshape(-1, 1) # Column vector
t2 = X2[:, 0].reshape(1, -1) # Row vector
t_min = np.minimum(t1, t2)
t_max = np.maximum(t1, t2)
# Compute the covariance matrix using the given formula
cov_matrix = self.sigma ** 2 * (t_min / 2) * (1 - t_min / (3 * t_max))
return cov_matrix
def Kdiag(self, X):
t = X[:, 0]
# Compute the diagonal elements of the covariance matrix
cov_diag = self.sigma ** 2 * (t / 2) * (1 - t / (3 * t))
# Simplify the expression
cov_diag = self.sigma ** 2 * (t / 2) * (1 - 1 / 3)
cov_diag = self.sigma ** 2 * (t / 2) * (2 / 3)
cov_diag = self.sigma ** 2 * t * (1 / 3)
return cov_diag
def update_gradients_full(self, dL_dK, X, X2=None):
pass
def gradients_X(self, dL_dK, X, X2=None):
return np.zeros(X.shape)
# Custom mean function to include drift
class TimeAverageMean(GPy.core.Mapping):
def __init__(self, input_dim, output_dim, S0=0.0, mu=0.0):
super(TimeAverageMean, self).__init__(input_dim, output_dim)
self.S0 = S0 # Initial value
self.mu = mu # Drift coefficient
def f(self, X):
t = X[:, 0]
mean = self.S0 + 0.5 * self.mu * t
return mean.reshape(-1, 1)
def update_gradients(self, dL_df, X):
pass
def gradients_X(self, dL_df, X):
return np.zeros(X.shape)
# Instantiate the Time Average kernel with parameters
ta_kernel = TimeAverageKernel(input_dim=1, sigma=sigma)
# Instantiate the mean function with S0 and mu
mean_function = TimeAverageMean(input_dim=1, output_dim=1, S0=S0, mu=mu)
# Provide the initial observation at t=1e-6 (approaching zero)
t_data = np.array([[1e-6]])
Y_data = mean_function.f(t_data) # Mean at t=1e-6
# Create a GP model with the custom mean function and Time Average kernel
model = GPy.models.GPRegression(t_data, Y_data, kernel=ta_kernel, mean_function=mean_function)
# Set noise variance to a negligible value since we're simulating without observation noise
model.likelihood.variance.fix(1e-10)
# Sample paths from the GP model
num_samples = 5
samples = model.posterior_samples_f(t, size=num_samples)
# Plot the sampled paths
plt.figure(figsize=(12, 6))
for i in range(num_samples):
plt.plot(t, samples[:, :, i], label=f'Path {i+1}')
plt.title('Sampled Paths of the arithmetic mean of a Brownian motion')
plt.xlabel('Time t')
plt.ylabel('M(t)')
plt.grid(True)
plt.legend()
plt.show()
Geometric Brownian Motion#
Simulation using discrete paths#
import numpy as np
import matplotlib.pyplot as plt
def simulate_gbm(S0, mu, sigma, T, N, num_paths):
dt = T / N
t = np.linspace(0, T, N)
paths = []
for _ in range(num_paths):
W = np.random.normal(0, np.sqrt(dt), N)
W = np.cumsum(W) # cumulative sum to represent Brownian motion
S = S0 * np.exp((mu - 0.5 * sigma**2) * t + sigma * W)
paths.append(S)
return t, paths
# Parameters
S0 = 100 # Initial value of the asset
mu = 0.05 # Drift
sigma = 0.2 # Volatility
T = 1 # Time horizon (1 year)
N = 1000 # Number of time steps
num_paths = 5 # Number of paths to simulate
t, paths = simulate_gbm(S0, mu, sigma, T, N, num_paths)
# Plotting the simulated GBM paths
plt.figure(figsize=(10, 6))
for i, S in enumerate(paths):
plt.plot(t, S, label=f'Path {i + 1}')
plt.xlabel('Time')
plt.ylabel('Price')
plt.title('Sampled paths from Geometric Brownian Motion')
plt.grid(True)
plt.show()
Simulation using Gaussian Processes#
import GPy
import numpy as np
import matplotlib.pyplot as plt
# Parameters for Geometric Brownian Motion
S0 = 100 # Initial value of the asset
mu = 0.05 # Drift
sigma = 0.2 # Volatility
T = 1 # Time horizon (1 year)
n = 1000 # Number of steps
t = np.linspace(0, T, n+1).reshape(-1, 1) # Time points
# Define the custom kernel for Brownian motion
class GBMKernel(GPy.kern.Kern):
def __init__(self, input_dim, sigma=1.0, active_dims=None):
super(GBMKernel, self).__init__(input_dim, active_dims, 'Brownian')
self.sigma = GPy.core.Param('sigma', sigma)
self.link_parameters(self.sigma)
def K(self, X, X2=None):
if X2 is None:
X2 = X
t1 = X
t2 = X2.T
# Covariance function of Brownian motion: sigma^2 * min(s, t)
cov_matrix = self.sigma**2 * np.minimum(t1, t2)
return cov_matrix
def Kdiag(self, X):
return self.sigma**2 * X[:, 0]
# No-op for gradient update since we are not optimizing the kernel
def update_gradients_full(self, dL_dK, X, X2=None):
pass
# No-op for kernel gradient since we are not optimizing the kernel
def gradients_X(self, dL_dK, X, X2=None):
return np.zeros(X.shape)
# Custom mean function to include drift
class GBMMean(GPy.core.Mapping):
def __init__(self, input_dim, output_dim, S0=0.0, mu=0.0):
super(GBMMean, self).__init__(input_dim, output_dim)
self.S0 = S0 # Initial value
self.mu = mu # Drift coefficient
def f(self, X):
# Mean function: S0 + mu * t
return self.S0 + self.mu * X
def update_gradients(self, dL_df, X):
pass
def gradients_X(self, dL_df, X):
return np.zeros(X.shape)
# Instantiate the Brownian motion kernel with parameters
bm_kernel = GBMKernel(input_dim=1, sigma=sigma)
# Instantiate the mean function with S0 and mu
mean_function = GBMMean(input_dim=1, output_dim=1, S0=np.log(S0), mu=mu)
# Provide the initial observation at t=0
t_data = np.array([[0.0]])
Y_data = np.array([[np.log(S0)]])
# Create a GP model with the custom mean function and Brownian motion kernel
model = GPy.models.GPRegression(t_data, Y_data, kernel=bm_kernel, mean_function=mean_function)
# Set noise variance to a negligible value since we're simulating without observation noise
model.likelihood.variance.fix(1e-10)
# Sample paths from the GP model
num_samples = 5
samples = model.posterior_samples_f(t, size=num_samples)
# Transform samples to original GBM scale
gbm_samples = np.exp(samples)
# Plot the sampled paths
plt.figure(figsize=(10, 6))
for i in range(num_samples):
plt.plot(t, gbm_samples[:, :, i], label=f'Path {i+1}')
plt.title(f'Sampled Paths from Geometric Brownian Motion')
plt.xlabel('Time t')
plt.ylabel('S(t)')
plt.grid(True)
plt.legend()
plt.show()
Orstein - Uhlenbeck process#
Simulation using discrete paths#
import numpy as np
import matplotlib.pyplot as plt
# Parameters for the Ornstein-Uhlenbeck process
S0 = 5.0 # Initial value
mu = 1.0 # Long-term mean
theta = 10 # Rate of mean reversion
sigma = 1 # Volatility
T = 1.0 # Total time
n = 100 # Number of steps
dt = T / n # Time step size
t = np.linspace(0, T, n+1) # Time points
# Number of paths to simulate
num_paths = 5
# Set up the plot
plt.figure(figsize=(10, 6))
# Simulate multiple paths
for _ in range(num_paths):
S = np.zeros(n+1) # Initialize the process
S[0] = S0 # Set initial value
for i in range(1, n+1):
# Compute the mean and variance for the next step
mean = S0 * np.exp(-theta * t[i]) + mu * (1 - np.exp(-theta * t[i]))
variance = (sigma**2 / (2 * theta)) * (1 - np.exp(-2 * theta * t[i]))
# Sample from the normal distribution with the computed mean and variance
S[i] = np.random.normal(mean, np.sqrt(variance))
# Plot the Ornstein-Uhlenbeck path
plt.plot(t, S, label=f'Path {_+1}')
# Customize the plot
plt.title('Sampled Paths from the Ornstein-Uhlenbeck Process')
plt.xlabel('Time t')
plt.ylabel('S(t)')
plt.grid(True)
plt.legend()
plt.show()
Simulation using Gaussian Processes#
import GPy
import numpy as np
import matplotlib.pyplot as plt
# Parameters for the Ornstein-Uhlenbeck process
S0 = 5.0 # Initial value
mu = 1.0 # Long-term mean
theta = 10 # Rate of mean reversion
sigma = 1 # Volatility
T = 1.0 # Total time
n = 1000 # Number of steps
t = np.linspace(0, T, n+1).reshape(-1, 1) # Time points
# Define the custom kernel for the Ornstein-Uhlenbeck process
class OrnsteinUhlenbeckKernel(GPy.kern.Kern):
def __init__(self, input_dim, theta=1.0, sigma=1.0, active_dims=None):
super(OrnsteinUhlenbeckKernel, self).__init__(input_dim, active_dims, 'OU')
self.theta = GPy.core.Param('theta', theta)
self.sigma = GPy.core.Param('sigma', sigma)
self.link_parameters(self.theta, self.sigma)
def K(self, X, X2=None):
if X2 is None:
X2 = X
t1 = X
t2 = X2.T
min_t = np.minimum(t1, t2)
# Covariance function of the Ornstein-Uhlenbeck process
cov_matrix = (self.sigma**2 / (2 * self.theta)) * np.exp(-self.theta * np.abs(t1 - t2)) * (1 - np.exp(-2 * self.theta * min_t))
return cov_matrix
def Kdiag(self, X):
return (self.sigma**2 / (2 * self.theta)) * (1 - np.exp(-2 * self.theta * X[:, 0]))
# No-op for gradient update since we are not optimizing the kernel
def update_gradients_full(self, dL_dK, X, X2=None):
pass
# No-op for kernel gradient since we are not optimizing the kernel
def gradients_X(self, dL_dK, X, X2=None):
return np.zeros(X.shape)
# Custom mean function to include mean-reverting drift toward mu
class OUProcessMean(GPy.core.Mapping):
def __init__(self, input_dim, output_dim, S0=0.0, mu=0.0, theta=1.0):
super(OUProcessMean, self).__init__(input_dim, output_dim)
self.S0 = S0 # Initial value
self.mu = mu # Long-term mean
self.theta = theta # Rate of mean reversion
def f(self, X):
# Mean function: S_0 * exp(-theta * t) + mu * (1 - exp(-theta * t))
return self.S0 * np.exp(-self.theta * X) + self.mu * (1 - np.exp(-self.theta * X))
def update_gradients(self, dL_df, X):
pass
def gradients_X(self, dL_df, X):
return np.zeros(X.shape)
# Instantiate the OU kernel with parameters
ou_kernel = OrnsteinUhlenbeckKernel(input_dim=1, theta=theta, sigma=sigma)
# Instantiate the mean function with non-zero initial value S0 and long-term mean mu
mean_function = OUProcessMean(input_dim=1, output_dim=1, S0=S0, mu=mu, theta=theta)
# Provide the initial observation at t=0
t_data = np.array([[0.0]])
Y_data = np.array([[S0]])
# Create a GP model with the custom mean function and OU kernel
model = GPy.models.GPRegression(t_data, Y_data, kernel=ou_kernel, mean_function=mean_function)
# Set noise variance to a negligible value since we're simulating without observation noise
model.likelihood.variance.fix(1e-10)
# Sample paths from the GP model
num_samples = 5
samples = model.posterior_samples_f(t, size=num_samples)
# Plot the sampled paths
plt.figure(figsize=(10, 6))
for i in range(num_samples):
plt.plot(t, samples[:, :, i], label=f'Path {i+1}')
plt.title(f'Sampled Paths from Ornstein-Uhlenbeck Process (S0={S0}, mu={mu}, theta={theta})')
plt.xlabel('Time t')
plt.ylabel('S(t)')
plt.grid(True)
plt.legend()
plt.show()
Brownian bridge#
Simulation using discrete paths#
import numpy as np
import matplotlib.pyplot as plt
# Parameters for the Brownian bridge
T = 1.0 # Total time
n = 100 # Number of steps
dt = T / n # Time step size
t = np.linspace(0, T, n+1) # Time points
# Number of paths to simulate
num_paths = 5
# Set up the plot
plt.figure(figsize=(10, 6))
# Simulate multiple paths
for _ in range(num_paths):
W = np.zeros(n+1) # Initialize the Wiener process
for i in range(1, n+1):
W[i] = W[i-1] + np.sqrt(dt) * np.random.randn() # Standard Wiener process
# Adjust Wiener process to form a Brownian bridge
B = W - (t / T) * W[-1] # Brownian bridge: W(t) - (t/T) * W(T)
# Plot the Brownian bridge path
plt.plot(t, B, label=f'Path {_+1}')
# Customize the plot
plt.title('Sampled Paths from a Brownian Bridge (Discrete Time Simulation)')
plt.xlabel('Time t')
plt.ylabel('B(t)')
plt.grid(True)
plt.legend()
plt.show()
Simulation using Gaussian processes#
import GPy
import numpy as np
import matplotlib.pyplot as plt
# Time points at which to sample the Brownian bridge
T = 1.0
t = np.linspace(0, T, 100).reshape(-1, 1)
# Define a custom kernel for the Brownian bridge
class BrownianBridgeKernel(GPy.kern.Kern):
def __init__(self, input_dim, variance=1.0, active_dims=None):
super(BrownianBridgeKernel, self).__init__(input_dim, active_dims, 'brownian_bridge')
self.variance = GPy.core.Param('variance', variance)
self.link_parameter(self.variance)
def K(self, X, X2=None):
if X2 is None:
X2 = X
t1 = X
t2 = X2.T
T = np.max(X)
# Brownian bridge kernel: min(t1, t2) - (t1 * t2) / T
cov_matrix = np.minimum(t1, t2) - (t1 * t2) / T
return self.variance * cov_matrix
def Kdiag(self, X):
return self.variance * (X[:, 0] - (X[:, 0]**2) / T)
# No-op for gradient update since we are not optimizing the kernel
def update_gradients_full(self, dL_dK, X, X2=None):
pass
# No-op for kernel gradient since we are not optimizing the kernel
def gradients_X(self, dL_dK, X, X2=None):
return np.zeros(X.shape)
# Instantiate the Brownian bridge kernel with variance 1
brownian_bridge_kernel = BrownianBridgeKernel(input_dim=1, variance=1.0)
# Create a GP model with zero mean function
mean_function = GPy.mappings.Constant(input_dim=1, output_dim=1, value=0)
model = GPy.models.GPRegression(t, np.zeros_like(t), kernel=brownian_bridge_kernel, mean_function=mean_function)
# Sample paths from the GP model without optimizing (since Brownian bridge doesn't require fitting)
num_samples = 5
samples = model.posterior_samples_f(t, size=num_samples)
# Plot the sampled paths
plt.figure(figsize=(10, 6))
for i in range(num_samples):
plt.plot(t, samples[:, :, i], label=f'Sample {i+1}')
plt.title('Sampled Paths from a Brownian Bridge using Gaussian Processes')
plt.xlabel('Time')
plt.ylabel('B(t)')
plt.grid(True)
plt.legend()
plt.show()
Jump processes#
Homogeneous Poisson process simulation#
import numpy as np
import matplotlib.pyplot as plt
# Parameters
lambda_rate = 0.2 # The rate parameter λ of the Poisson process
T = 50 # Total time
delta_t = 0.01 # Time-step Δt
num_steps = int(T / delta_t) # Number of time steps
# Time grid
t = np.linspace(0, T, num_steps + 1)
# Initialize N_t
N_t = np.zeros(num_steps + 1, dtype=int)
# Simulate the Poisson process
for i in range(1, num_steps + 1):
# Generate a random number between 0 and 1
u = np.random.rand()
# If u < λΔt, increment N_t
if u < lambda_rate * delta_t:
N_t[i] = N_t[i - 1] + 1
else:
N_t[i] = N_t[i - 1]
# Plot the Poisson process
plt.step(t, N_t, where='post')
plt.xlabel('Time t')
plt.ylabel('N(t)')
plt.title('Simulated Poisson Process')
plt.grid(True)
plt.show()
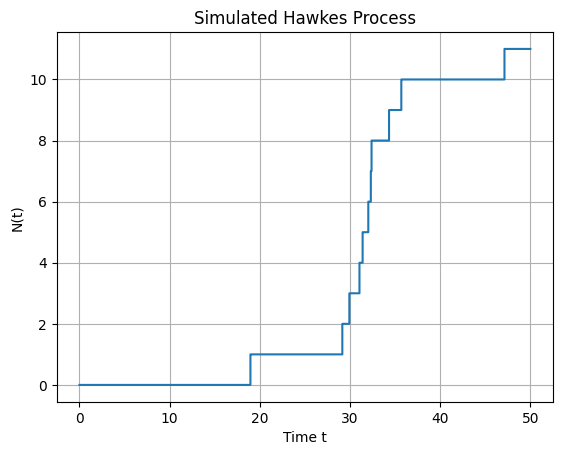
Hawkes process#
import numpy as np
import matplotlib.pyplot as plt
# Parameters
mu = 0.2 # Baseline intensity (μ)
alpha = 0.5 # Excitation parameter (α)
beta = 1 # Decay rate (β)
T = 50 # Total time to simulate
delta_t = 0.01 # Time-step Δt
# Time grid
time_grid = np.arange(0, T + delta_t, delta_t)
num_steps = len(time_grid)
# Initialize N_t and event times
N_t = np.zeros(num_steps, dtype=int)
event_times = []
# Simulate the Hawkes process
for i in range(1, num_steps):
t_i = time_grid[i]
# Compute lambda_t
lambda_t = mu
if event_times:
tau_array = np.array(event_times)
lambda_t += np.sum(alpha * np.exp(-beta * (t_i - tau_array)))
# Calculate the event probability
p = lambda_t * delta_t
if p > 1.0:
p = 1.0 # Ensure probability does not exceed 1
# Simulate event occurrence
u = np.random.rand()
if u < p:
N_t[i] = N_t[i - 1] + 1
event_times.append(t_i)
else:
N_t[i] = N_t[i - 1]
# Plot the counting process
#plt.figure(figsize=(12, 6))
plt.step(time_grid, N_t, where='post')
plt.xlabel('Time t')
plt.ylabel('N(t)')
plt.title('Simulated Hawkes Process')
plt.grid(True)
plt.show()
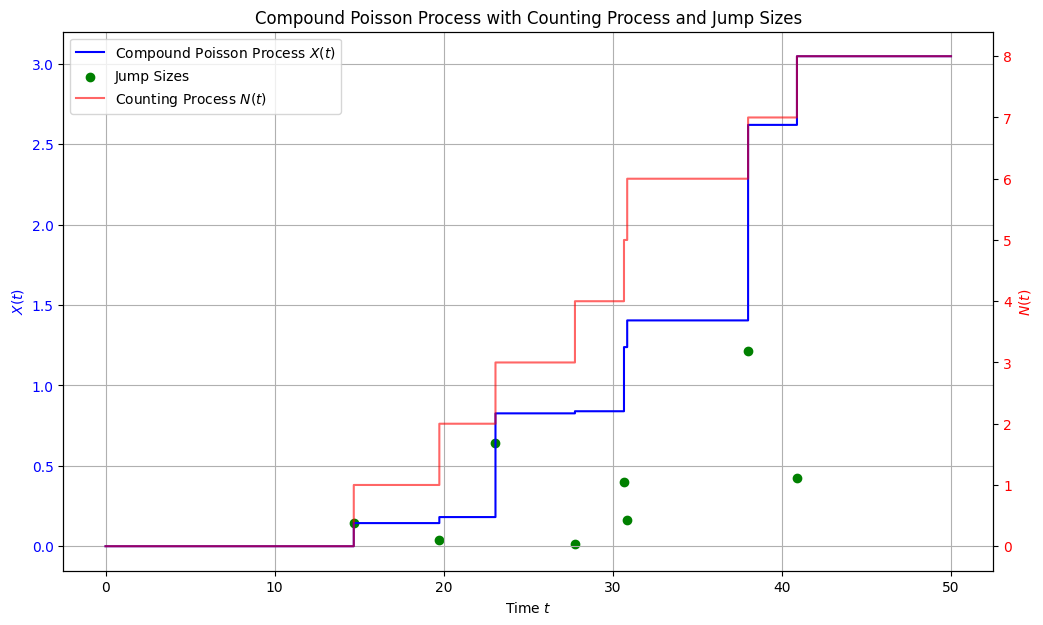
Compound Poisson process#
import numpy as np
import matplotlib.pyplot as plt
# Parameters
lambda_rate = 0.2 # Rate parameter λ of the Poisson process
T = 50 # Total simulation time
delta_t = 0.01 # Time-step Δt
num_steps = int(T / delta_t)
time_grid = np.linspace(0, T, num_steps + 1)
# Jump size distribution parameters
eta = 2.5 # Rate parameter of the exponential distribution for jump sizes
# Initialize arrays
N_t = np.zeros(num_steps + 1, dtype=int) # Counting process N(t)
X_t = np.zeros(num_steps + 1) # Compound Poisson process X(t)
event_times = [] # Times when events occur
jump_sizes = [] # Sizes of the jumps
# Simulate the compound Poisson process
for i in range(1, num_steps + 1):
t = time_grid[i]
# Probability of an event in [t_i, t_i + delta_t)
p = lambda_rate * delta_t
# Ensure probability does not exceed 1
p = min(p, 1.0)
# Determine if an event occurs
if np.random.rand() < p:
N_t[i] = N_t[i - 1] + 1
# Sample a jump size Y_i from an exponential distribution
Y_i = np.random.exponential(scale=1/eta)
jump_sizes.append(Y_i)
event_times.append(t)
X_t[i] = X_t[i - 1] + Y_i
else:
N_t[i] = N_t[i - 1]
X_t[i] = X_t[i - 1]
# Plot all three visualizations in one figure
fig, ax1 = plt.subplots(figsize=(12, 7))
# Plot the compound Poisson process X(t)
ax1.step(time_grid, X_t, where='post', label='Compound Poisson Process $X(t)$', color='blue')
ax1.set_xlabel('Time $t$')
ax1.set_ylabel('$X(t)$', color='blue')
ax1.tick_params(axis='y', labelcolor='blue')
ax1.grid(True)
# Plot jump sizes as scatter points on X(t)
event_indices = [np.searchsorted(time_grid, t) for t in event_times]
event_values = [X_t[idx] for idx in event_indices]
ax1.scatter(event_times, jump_sizes, color='green', marker='o', label='Jump Sizes')
# Create a secondary y-axis for the counting process N(t)
ax2 = ax1.twinx()
ax2.step(time_grid, N_t, where='post', label='Counting Process $N(t)$', color='red', alpha=0.6)
ax2.set_ylabel('$N(t)$', color='red')
ax2.tick_params(axis='y', labelcolor='red')
# Combine legends from both axes
lines1, labels1 = ax1.get_legend_handles_labels()
lines2, labels2 = ax2.get_legend_handles_labels()
ax1.legend(lines1 + lines2, labels1 + labels2, loc='upper left')
plt.title('Compound Poisson Process with Counting Process and Jump Sizes')
plt.show()
Jump diffusion processes#
import numpy as np
import matplotlib.pyplot as plt
def simulate_merton_jump_diffusion(S0, mu, sigma, lamb, mu_j, sigma_j, T, N, M):
"""
Simulate the Merton jump diffusion model.
Parameters:
- S0: Initial asset price.
- mu: Expected return (drift rate).
- sigma: Volatility of the continuous component.
- lamb: Jump intensity (average number of jumps per unit time).
- mu_j: Mean of the jump size distribution (of ln(J)).
- sigma_j: Standard deviation of the jump size distribution (of ln(J)).
- T: Total time horizon.
- N: Number of time steps.
- M: Number of simulation paths.
Returns:
- t: Array of time points.
- S: Matrix of simulated asset prices (M paths x N+1 time points).
"""
dt = T / N # Time step size
t = np.linspace(0, T, N + 1) # Time grid
# Precompute constants
drift = (mu - 0.5 * sigma**2) * dt
diffusion = sigma * np.sqrt(dt)
# Initialize asset price paths
S = np.zeros((M, N + 1))
S[:, 0] = S0
for m in range(M):
# Initialize variables for each path
S_m = np.zeros(N + 1)
S_m[0] = S0
# Generate random numbers for Brownian motion
Z = np.random.normal(size=N)
# Generate random numbers for jumps
# Poisson random numbers for number of jumps in each time step
Nt = np.random.poisson(lamb * dt, size=N)
# Total number of jumps
num_jumps = np.sum(Nt)
# Generate jump sizes only where jumps occur
if num_jumps > 0:
jump_sizes = np.random.normal(loc=mu_j, scale=sigma_j, size=num_jumps)
else:
jump_sizes = np.array([])
# Index to keep track of jump sizes
jump_index = 0
for i in range(1, N + 1):
# Continuous part
S_m[i] = S_m[i - 1] * np.exp(drift + diffusion * Z[i - 1])
# Jump part
if Nt[i - 1] > 0:
# Apply jumps
total_jump = np.sum(jump_sizes[jump_index:jump_index + Nt[i - 1]])
S_m[i] *= np.exp(total_jump)
jump_index += Nt[i - 1]
S[m, :] = S_m
return t, S
# Parameters
S0 = 100 # Initial asset price
mu = 0.1 # Expected return
sigma = 0.2 # Volatility
lamb = 1 # Jump intensity λ
k = -0.1 # Mean of ln(J) (jump sizes)
delta = 0.1 # Std dev of ln(J)
T = 1.0 # Total time (e.g., 1 year)
N = 1000 # Number of time steps
M = 5 # Number of simulation paths
# Simulate the Merton jump diffusion model
t, S = simulate_merton_jump_diffusion(S0, mu, sigma, lamb, k, delta, T, N, M)
# Plot the simulated asset price paths
plt.figure(figsize=(12, 6))
for m in range(M):
plt.plot(t, S[m, :], label=f'Path {m + 1}')
plt.xlabel('Time t')
plt.ylabel('Asset Price S(t)')
plt.title('Merton Jump Diffusion Model Simulation')
plt.legend()
plt.grid(True)
plt.show()